
Professors Jason Adolf, Keith Dunton, and John Tiedemann are taking the Census. Clipboards in hand, they and a crew of student researchers stop by some of the Monmouth County Bayshore’s most diverse neighborhoods, collecting information that will help build a clearer picture of who lives there. On this day, we learn the area has high populations of Northern pufferfish, squid, butterfish, sea robins and spider crabs.
 The team also has a way to learn about those who weren’t seen at the time of the visit. Water samples taken at the scene will be lab tested for trace genetic material, known as environmental DNA (eDNA), that can identify marine organisms which may or may not have been caught in their nets.
The team also has a way to learn about those who weren’t seen at the time of the visit. Water samples taken at the scene will be lab tested for trace genetic material, known as environmental DNA (eDNA), that can identify marine organisms which may or may not have been caught in their nets.
“You ask the question, do the fish that I saw in the trawl match the fish that the DNA sequence turns up?” said Adolf, endowed associate professor of marine science at Monmouth University. “Do the phytoplankton that I saw in the microscope match the phytoplankton that the DNA sequences turn up? Do the benthic invertebrates that we caught in the grab sampler match what we saw in the DNA?”
 The answers will not only yield important information about marine life in Sandy Hook and Raritan bays, but for Monmouth University’s continuing exploration on the utility of eDNA as a tool for taking inventories of ecosystem fisheries and wildlife.
The answers will not only yield important information about marine life in Sandy Hook and Raritan bays, but for Monmouth University’s continuing exploration on the utility of eDNA as a tool for taking inventories of ecosystem fisheries and wildlife.
On Aug. 11, Adolf, students Skyler Post and Bryce McCall, Tiedemann, Dunton and UCI Marine Scientist Jim Nickels sampled six sites from the tip of Sandy Hook to the mouth of the Raritan River. Although only a few miles apart, the habitats at these stations can vary widely. The Lower Hudson-Raritan Estuary (to which Sandy Hook and Raritan bays belong) experiences a constant intermixing of saltwater, suburban rivers like the Navesink and Shrewsbury, industrially developed bodies such as the Arthur Kill and Hudson River, and numerous other New York-New Jersey tributaries. The floor at some stations are sandy, while others are muddy or rocky.
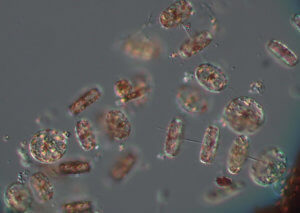
In addition to marine life data, the team records oceanographic parameters such as salinity, dissolved oxygen, pH, temperature, clarity, and phytoplankton biomass. Each can be indicators of the general health of the waters. Through the day’s sampling, the group learned of a large phytoplankton bloom centered along the transect between Raritan Bay and the tip of Sandy Hook, and hypoxic bottom waters (waters without sufficient oxygen to support most marine life) toward the western portion of the transect.
The water samples for eDNA analysis are run through filters, frozen and handed over to UCI Marine Genetics Faculty Fellow Megan Phifer-Rixey for analysis. This semester, she and students Cameron Gaines and Lilia Crew will extract the DNA and compare it to a database of genetic barcodes for other organisms, looking for matches.
“There’s usually quite a bit of DNA in the water samples we get from the trawls,” Phifer-Rixey said. “There’s a lot of life out there.”
 With grant funding from the Achelis & Bodman Foundation, the project’s principal investigators, Adolf, Dunton and Phifer-Rixey, will conduct a half-dozen research cruises through the fall of 2021 at the same locations, building a foundation of data that can be used to detect important trends over time. Adolf said he’d like to continue the work well beyond the grant period so Monmouth will one day have decades of this data to draw upon.
With grant funding from the Achelis & Bodman Foundation, the project’s principal investigators, Adolf, Dunton and Phifer-Rixey, will conduct a half-dozen research cruises through the fall of 2021 at the same locations, building a foundation of data that can be used to detect important trends over time. Adolf said he’d like to continue the work well beyond the grant period so Monmouth will one day have decades of this data to draw upon.
“A time series is like a garden. You start small, you hope for something, and over time you watch it produce more and more results,” Adolf said. “I’d like to do six cruises per year for the rest of my career at Monmouth. This generates the kind of data needed to understand the effects of climate and anthropogenic change on marine systems, and provides ongoing opportunities for Monmouth students to become involved in established research projects.”
Click here to view an album of photos from the cruise. For more on eDNA, see the proceedings of the National Conference on Marine Environmental DNA hosted by Rockefeller University Program for the Human Environment and the UCI.
